
I’m a data scientist at Princeton University, formerly in Computer Science and currently in Ecology and Evolutionary Biology. I’m also affiliated with the Center for Statistics and Machine Learning.
My interests and expertise are in bioinformatics, software engineering, machine learning, and analyzing large-scale datasets using extensive visualizations.
As a senior data scientist, I primarily collaborate as an independent contributor, rotating among labs (~4 month terms to maximize diversity) and queueing projects via networking within the department. Additionally, I:
- develop computational workflows that enable or accelerate several projects
- mentor individual students or postdocs
- teach workshops
- serve as consultant
Highlights
Here, I highlight my technical contributions to selected collaborations in which I take the lead on a modular component. These contributions involve applying my skills mentioned above to various data modalities including spatial transcriptomics, 3D movies of neural activity, and whole-genome sequencing (WGS).
- GASTON: deep neural network to segment domains and study continuous variation in gene expression from spatial transcriptomics data
- refactored code into python package to run at scale and optimize neural network architecture
- feature-ized histology images (H&E stained) to facilitate tissue segmentation
- analyzed several colorectal cancer datasets to characterize metabolic gradients and the tumor microenvironment
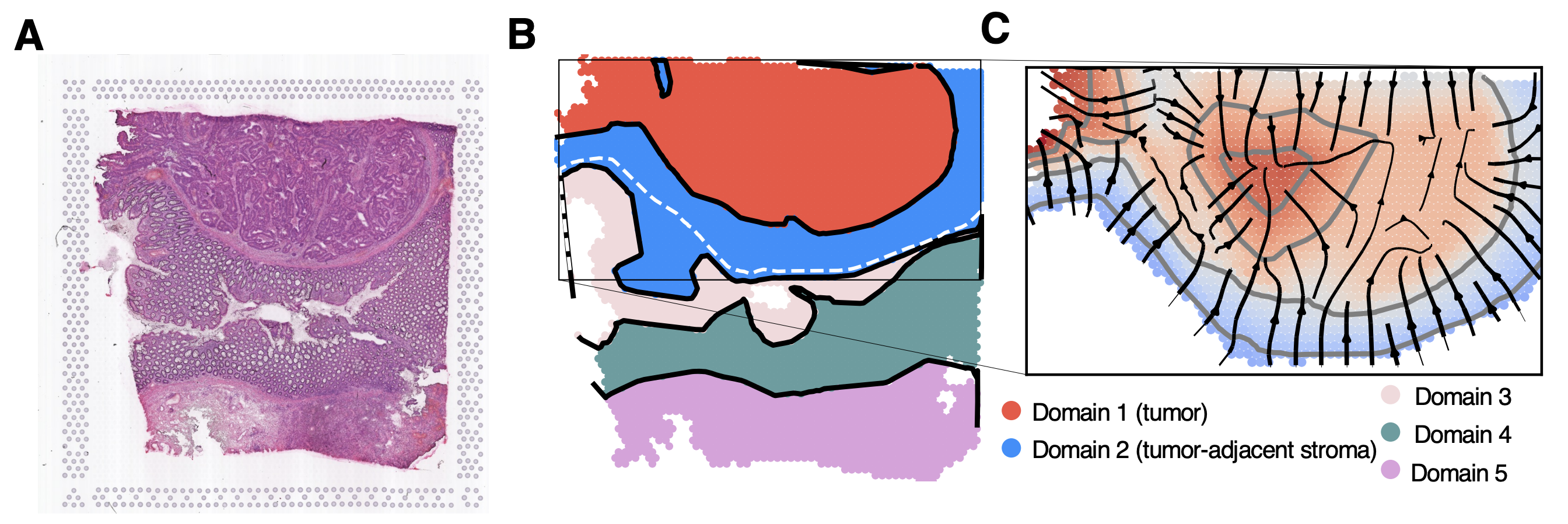
(A) H&E stain of a 10x Genomics Visium colorectal tumor sample. (B) Spatial domains learned by GASTON. Domains 1 and 2 are labeled as tumor and tumor-adjacent stroma, respectively, based on the histology image in (A). (C) Spatial gradients learned by GASTON show directions of maximum gene expression changes in tumor and tumor-adjacent stromal domains.
- 3D movie analysis: image segmentation to decode neural activity in the mosquito antennal lobe (AL)
- created python workflows to measure activity of the entire AL
- discovered technical batch effects and used the experimental design to correct them via custom statistical models
- segmented individual glomeruli (clusters of nerve endings) within the AL via nonnegative matrix factorization (NMF)
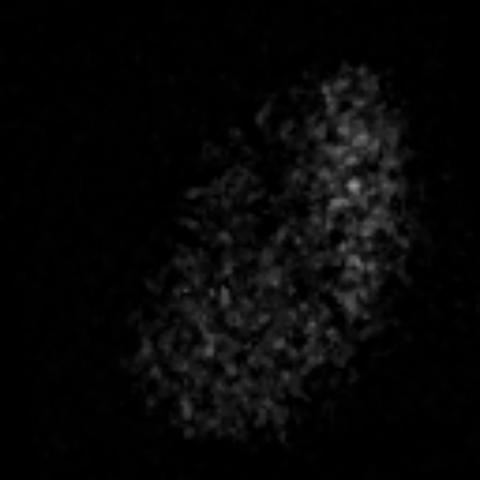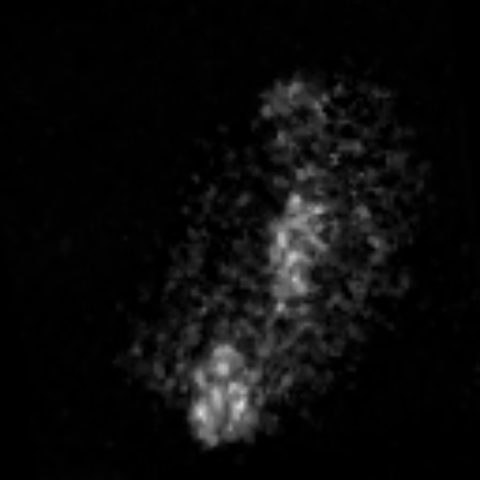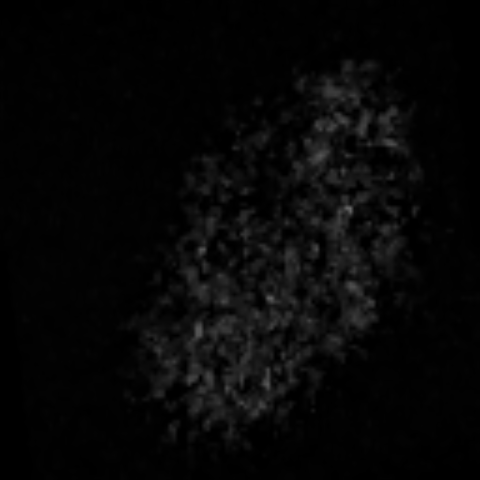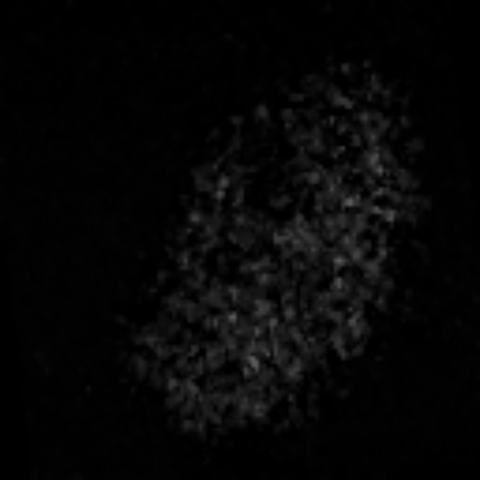
2D max projection of raw data from the AL of a mosquito that recieved an odor puff. These data are noisy even after median filtering.
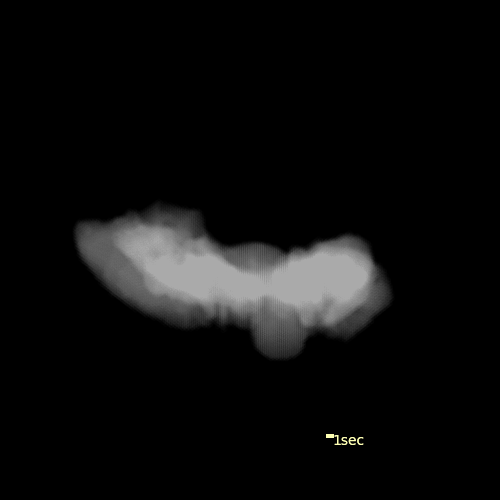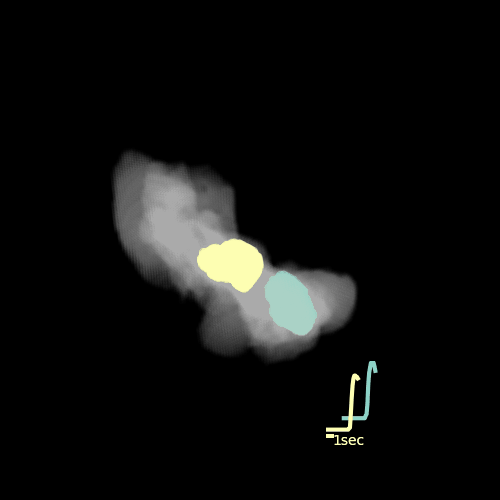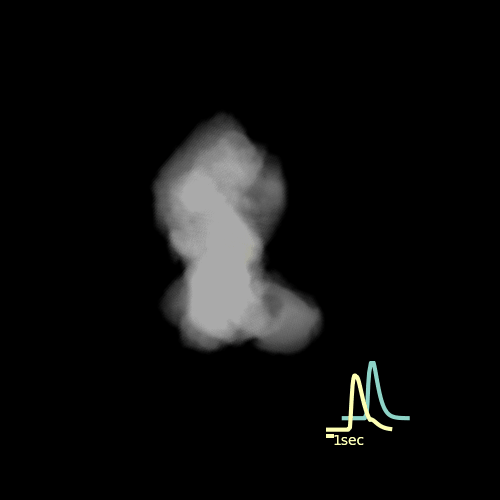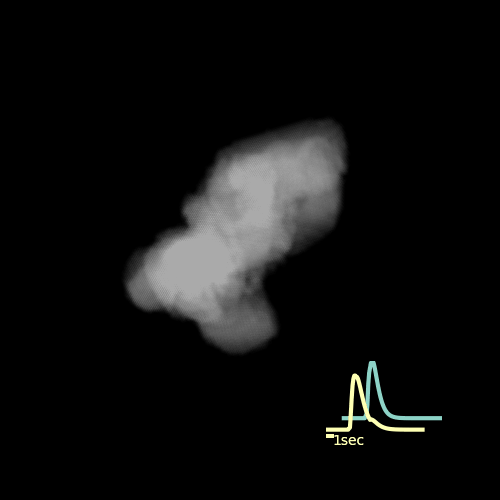
Custom visualization of NMF segmentation shows spatial positions and activity traces of glomeruli.
- HATCHet2: copy-number calling (amplifications/deletions of DNA) for tumor WGS data
- wrote several modules to phase genotypes into haplotypes using the 1000GP reference panel
- Docker-ized software for cloud computing on GCP and AWS
- analyzed large datasets in collaboration with the Genomic Data Analysis Network
- gave virtual invited talk at University of Edinburgh’s biomedical AI seminar
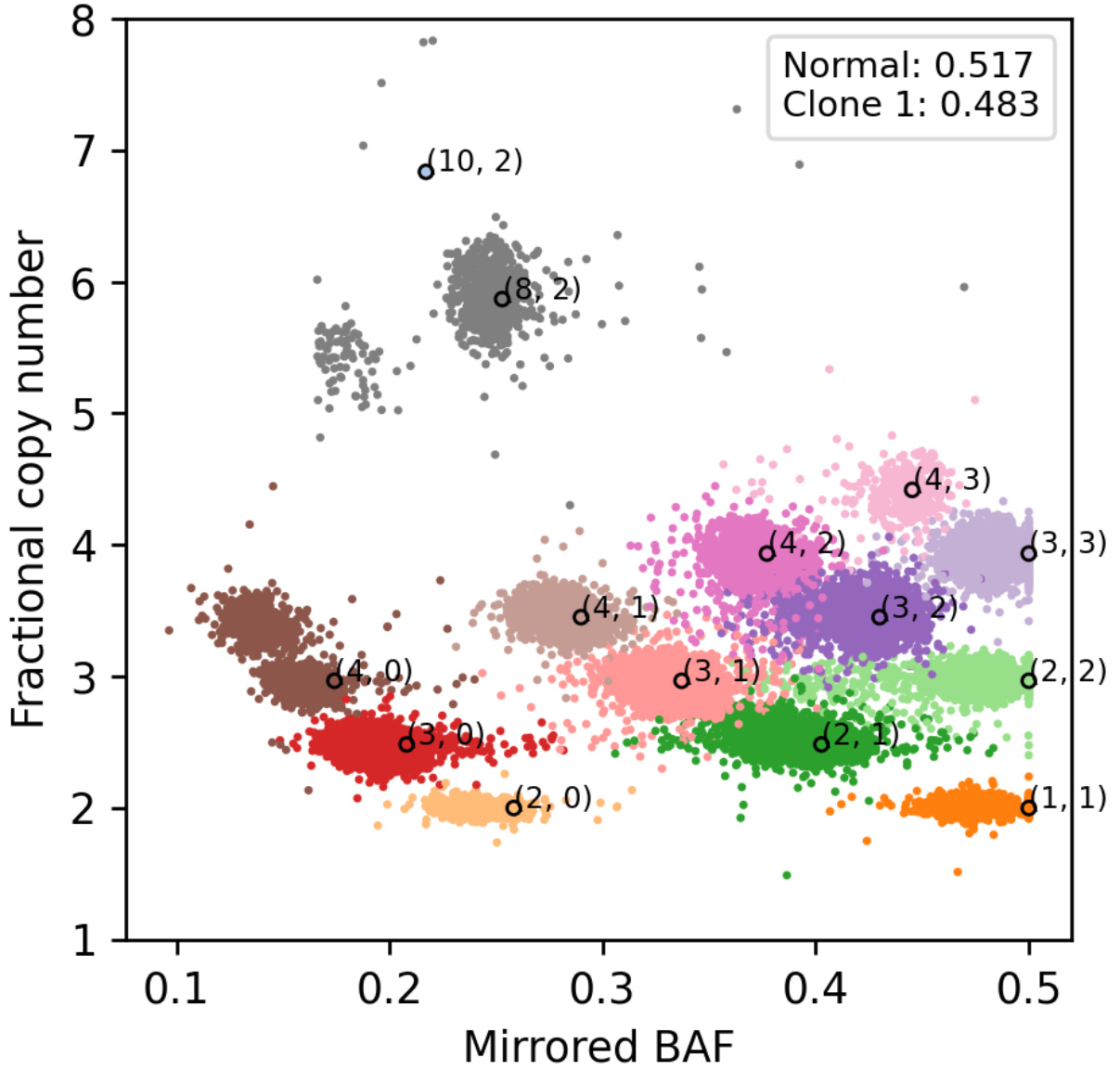
From tumor WGS data, HATCHet2 extracts 2 features that are correlated with copy number: the fractional copy number (rescaled from read depth) and the mirrored B-allele frequency (BAF). For normal diploids, these values should be 2 and 0.5, respectively, which we observe for some genomic regions in this sample (orange dots). However, most tumors exhibit stiking variation in copy-number along the genome.
- snpArcher: workflow to automate variant calling in nonmodel organisms
- authored original snakemake code and supplementary algorithms to massively parallelize two variant callers
- Tuskless African elephants: poaching drives evolution of tusklessness, a female-specific trait encoded by a male-lethal mutation
- contributed all genomic analyses as co-first author
- led to over 411 news stories from 301 media outlets
- invited guest on the Nice Genes! podcast